Manage quantum chemistry computations at scale
I'd like to do machine learning on dataset X from a single line.
I want to generate bespoke parameters for a new drug from an existing database.
Which DFT method gives the best results for my set of reactions?
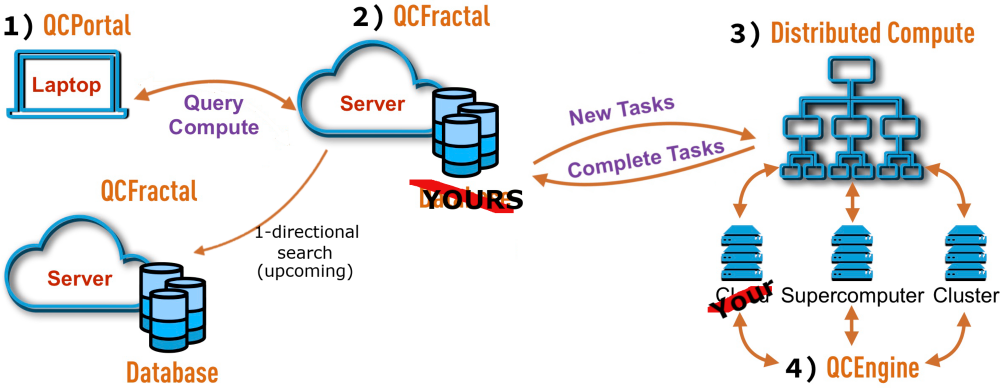
These are some of the use cases we at MolSSI heard from the quantum chemistry community when evaluating how best to serve this area. The emerging trend we saw was a need to have a regular structure of quantum chemistry results, the ability to access them on-demand without downloading several GB of data and run new calculations if need be ideally without repeating what someone has already done. To meet these goals, the QCArchive project was started to unite this otherwise isolated data which may only have been shared previously though massive, non-regular files; or through publications and supplementary material. Funded by the NSF, MolSSI seeks to provide solutions to these, and other use-cases in the quantum chemistry field through the QCArchive project.