Today, the Molecular Sciences Software Institute (MolSSI) is pleased to announce the Quantum Chemistry Archive (QCArchive) is officially entering stable beta status. The QCArchive project focuses on sharing, computing, and analyzing complex quantum chemistry data through a central server hosted by MolSSI for the entire Computational Molecular Science (CMS) community. In addition to this central server, the entire QCArchive software stack is open-source and can be used to store and evaluate data privately with the complete functionality of the MolSSI server. This infrastructure also provides several smaller modules which form the foundation of a modular software ecosystem. We originally announced QCArchive as an idea and a prototype on March 31st, 2019; since this time,we have progressed rapidly to provide the first release, ready for adoption by power users within the community.
Before we dive too deeply into what the stable beta means, let us first give an update on the main MolSSI QCArchive instance. In the past four months, we have computed, organized, and archived ~4.5M quantum chemical results with the help of our partners. In particular, the Open Force Field Initiative has evaluated 2,380 torsion drives, 169,000 geometry optimizations, and 50,000 Hessians for their force field fitting efforts. These sponsored calculations utilized multiple physical sites simultaneously through the MolSSI QCArchive central queue. The Sherrill group (Georgia Tech) has contributed over 30 gold-standard benchmark datasets from the literature, including S22, S66, and NHTBH38. MolSSI has further extended these datasets to include popular DFT functionals, Hartree–Fock, and MP2 computations in a variety of standard basis sets. We are also working with the de Jong group (LBNL) to evaluate thousands of metabolites with semiempirical methods. All of this data is currently freely accessible and compliant with FAIR data standards. You can get started with the QCArchive’s data via our Python front-end, QCPortal.
We anticipate that aggregating and disseminating data at this scale will be of immense use to the community. As we continue to gather data, we plan to simultaneously build a number of web applications to leverage this data towards common CMS research uses and undergraduate education. Hosted on qcarchive.molssi.org, these apps will allow browsing of the data natively in the browser – without a Python front end. A few web application ideas that we are either working on or planning:
- An interactive educational web portal where a simple search for e.g. “caffeine” will return a molecular geometry and plots of orbitals, dipoles, charges, ESP, and much more. This will query data from the MolSSI server without the need to recompute the data.
- View standard literature benchmark datasets such as S22 and GMTKN and compare the performance of DFT and other approximations on these benchmarks.
- A recommendation system for electronic structure methods. Given a set of molecules of interest, this ML-based app will suggest appropriate density functional or wavefunction methods.
If these ideas interest you or you have similar ideas you would like to build out, please do get in touch ! Our mission is to support researchers working on these ideas through rigorous reproducible data, high-quality software infrastructure, and intuitive user gateways.
Behind the QCArchive is a powerful modular open-source infrastructure:
- QCElemental - Periodic table information, version-controlled physical constants, molecule parsing, testing infrastructure, and MolSSI QCSchema models.
- QCEngine - Quantum chemistry program executor and IO standardizer (QCSchema).
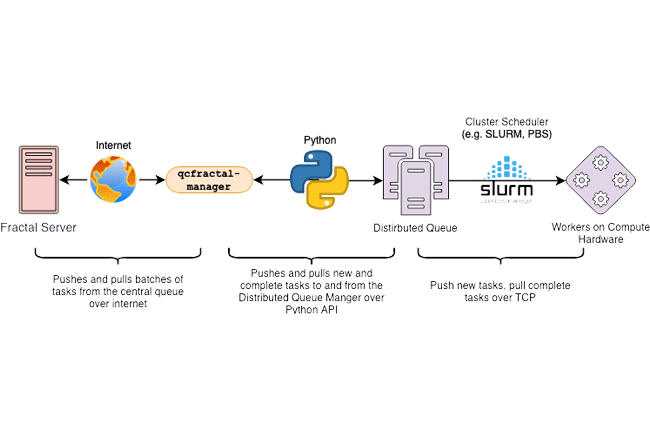
- QCFractal - Distributed task scheduler and executor, database store for chemistry results, and organization of results at scale.
- QCPortal - Visualization, organization, and statistical data for chemistry-related results and a front-end client for QCFractal.
These projects can be used independently as libraries in your project, or together to run a private instance of the QCArchive stack. We are currently seeing an uptake of the lower level libraries like QCElemental and QCEngine to power a variety of projects such as QUBEKit, Psi4, geomeTRIC, OptKing, and MultirefPredict.
This stable beta release means several things for the infrastructure:
- We are satisfied that the core infrastructure components are in a state that is extensible for the future without significant overhauls. Several extensive component overhauls are now complete and unlikely to change in the future.
- All data in QCFractal can be migrated forwards, and no data will be lost.
- The general API’s are stable, and any API-breaking changes will be following by deprecation warnings.
Early adoption of the infrastructure and tools is off to a great start, but we need help from the CMS community to make the QCArchive project even better. Reach out to us and become a partner or sponsor to help shape the future of the QCArchive. Let us know what features you want to see and how we can help with your use cases. Do you have specific calculations you want to run? Talk to us about leveraging our compute resources!
The MolSSI QCArchive project was created to support the computational molecular science community to help create, share, and understand data. We have begun to take the first steps in this journey and will continue to expand and refine these tools until they are useful to everyone in the community.
- The QCArchive Team